
- #TIDE GRAPH SEWELLS PT SOFTWARE#
- #TIDE GRAPH SEWELLS PT CODE#
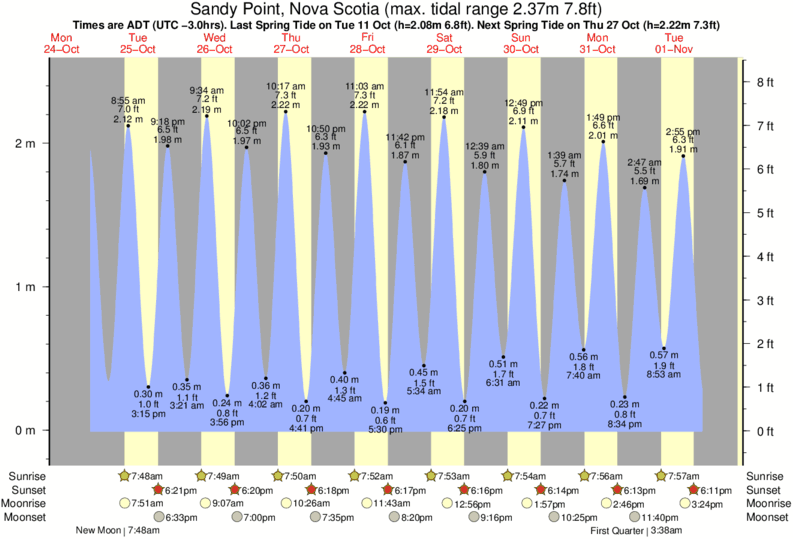
We advise to sequence a stretch of DNA ~700bp enclosing the designed editing site. Upload always the same amount of control as test samples. DNA of pool of cells treated with both Cas9 and the sgRNA).
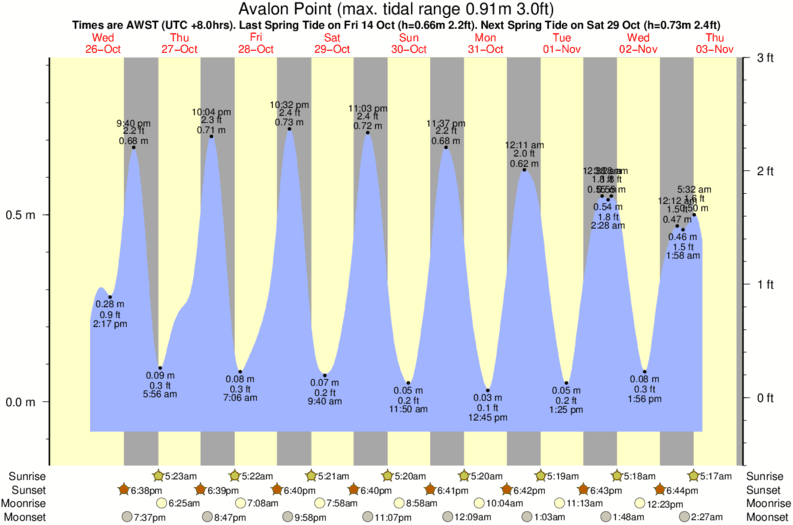
transfected without Cas9 or without the sgRNA) and the test samples (e.g. Upload the chromatogram sequence files of respectively the control samples (e.g.TIDE assumes that a dsDNA break is induced between nucleotides 17 and 18 in the sgRNA sequence. Numbers and other invalid (non-IUPAC) DNA characters will be automatically removed. Enter a 20nt ('5-'3) DNA character string representing the used sgRNA guides sequence immediately upstream of the PAM sequence (PAM not included).The Lawson-Hanson algorithm for non-negative least squares (NNLS). sangerseqR: Tools for Sanger Sequencing Data in R. Biostrings: String objects representingīiological sequences, and matching algorithms. R: A language and environment for statistical computing. For more information and to report bugs, please contact Acknowledgements This web tool was developed by Eva Brinkman, Tao Chen and Bas van Steensel from the Bas van Steensel lab. Stichting het Nederlands Kanker Instituut - Antoni van Leeuwenhoek ziekenhuis (The Netherlands Cancer Institute). Support is given by Data Curators VOF, see our.
#TIDE GRAPH SEWELLS PT CODE#
The R code of the TIDE algorithm is available upon request. #TIDE GRAPH SEWELLS PT SOFTWARE#
If you use this software for data analysis in a publication, please cite. The TIDE software is being provided as a free web service for research, educational, instructional and non-commercial purposes only. The output of TIDE is a comprehensive profile of all insertions and deletions (indels) in the edited sample. The input to TIDE is Sanger sequencing data. Based on the quantitative sequence trace data from two standard capillary sequencing reactions the TIDE software quantifies the editing efficacy and identifies the predominant types of insertions and deletions (indels) in the DNA of a targeted cell pool. TIDE provides rapid and reliable assessment of genome editing experiments of a target locus. Reference (please cite!): Brinkman et al, Nucl. Drawbacks: The method will not capture megabase long deletions that can originate by CRISPR/Cas9 induced DSB. What it needs: Two standard capillary sequencing reactions. For templated CRISPR/Cas9 experiments, use the TIDER web tool. If you have many samples, go to the TIDE batch site. When to use: Quantification of small indels. What it does: Estimates the spectrum and frequency of small insertions and deletions (indels) generated in a pool of cells by genome editing tools such as CRISPR/Cas9, TALENs and ZFNs. TIDE: Tracking of Indels by DEcomposition Purpose






 0 kommentar(er)
0 kommentar(er)
